Welcome to TLSEA
An online tool for lncRNA set Enrichment Analysis
Enrichment analysis of LncRNAs is a key step in predicting the list of differentially expressed genes and disease association, because if operated correctly, it can reduce the cost of experimental verification. Therefore, the development of electronic methods for enrichment analysis has become a competitive research field.
TLSEA is a web server for enrichment analysis of LncRNAs. It is based on heterogeneous network data fusion, network embedding and integrated learning. It can provide the following functions.
1) Enrichment analysis without expansion. Enter a list of differentially expressed lncrnas for enrichment analysis.
2) Enrichment analysis with expansion. Input a list of differentially expressed lncrnas and select a similarity coefficient for enrichment analysis.
All analyses are performed at the levels of heterogeneous network topology. The full prediction datasets can also be downloaded in the "Download" page.
Comprehensive usage instructions can be found in the "Help" menu.
News
Jan 2022: the original TLSEA program is released.
Jul 2022: a stable enrichment analysis module has been established.
TLSEA works on Gene Symbol, not anything else.
Contact
Dr. Jianwei Li, school of artificial intelligence, Hebei University of Technology, Tianjin 300401, China
Email: lijianwei@hebut.edu.cn
This website has been tested by using Chrome, Microsoft Edge and Firefox browsers. Microsoft IE may not work well.
Citation: Jianwei Li, Zhiguang Li, Yinfei Wang, Hongxin Lin, Baoqin Wu. TLSEA:a tool of lncRNA set enrichment analysis based on multi-source heterogeneous information fusion.
Download the Lncrna similarity matrix for TLSEA here:lncRNA_similarity.
TLSEA Tutorial
How to run Enrichment analysis
- User input differential expression list.In addition, you can analyze the test data set by clicking the sample button
- Tlsea can enrich and analyze the differential expression list submitted by users. Two enrichment analysis methods are provided here: one is to perform enrichment analysis only on the user submitted list (the similarity coefficient is none); The other is to expand the list submitted by users by random walk, and then conduct enrichment analysis.
- Submit the request by clicking the "Run" button.
How to interpret enrichment analysis results
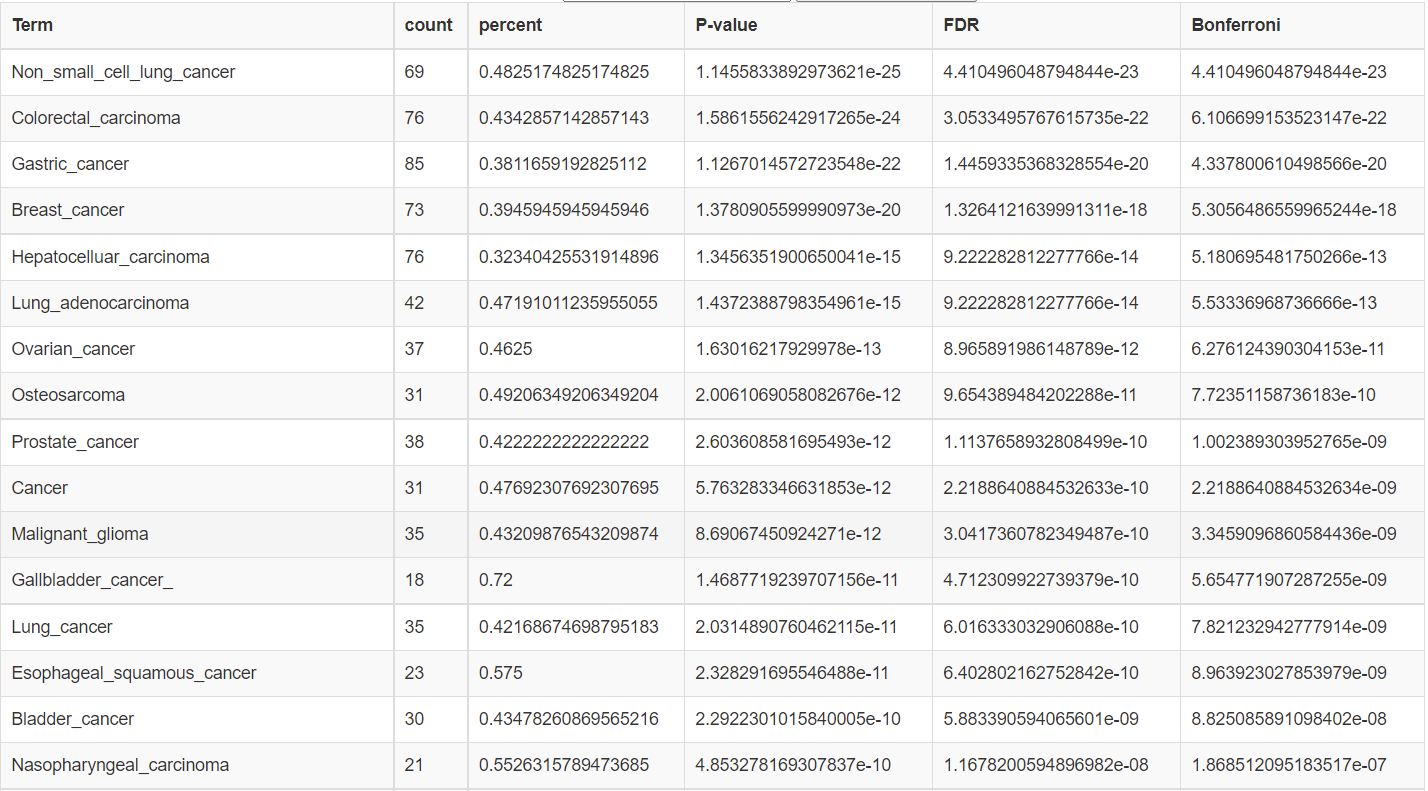
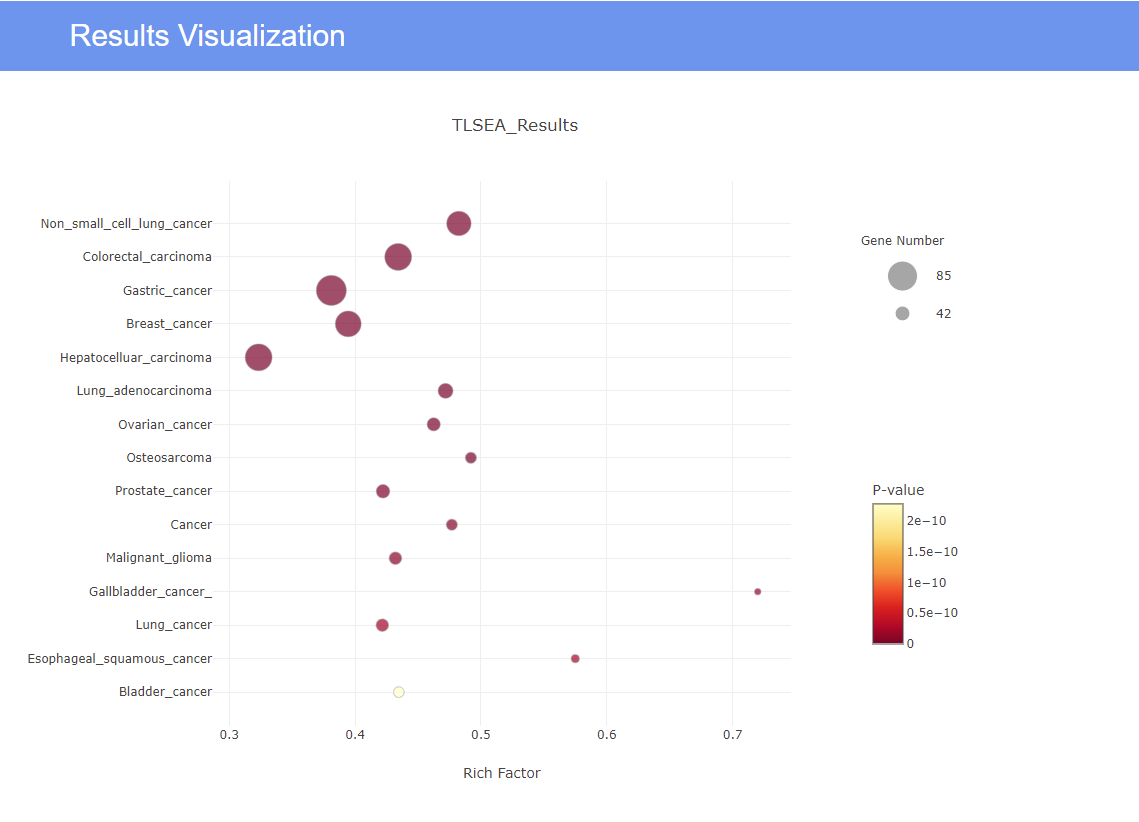
- Results of enrichment analysis. Arrange the order according to the p value and display the Term, count, percent, P-value, FDR and Bonferroni with P-value less than 0.01.
- If the similarity coefficient is selected, the expanded list will be displayed additionally.In addition, download function is provided
- According to the input value, a bubble diagram with significant enrichment is generated. (the default is all diseases with P < 0.01)
Download
The entire lncRNA similarity of TLSEA can be found on the download page, user can click 'lncRNA_similarity' link to download it.
