

Note that this page accepts human miRNA genes (pre-miRNAs) as its input. If the mature miRNA names (e.g. hsa-miR-140-5p) are provided, it will be collapsed into the corresponding miRNA gene (hsa-mir-140). If the exact name of the duplicated miRNA gene is not specified (e.g. hsa-mir-24), it will be mapped to all of the duplicated miRNA genes (hsa-mir-24-1 and hsa-mir-24-2).
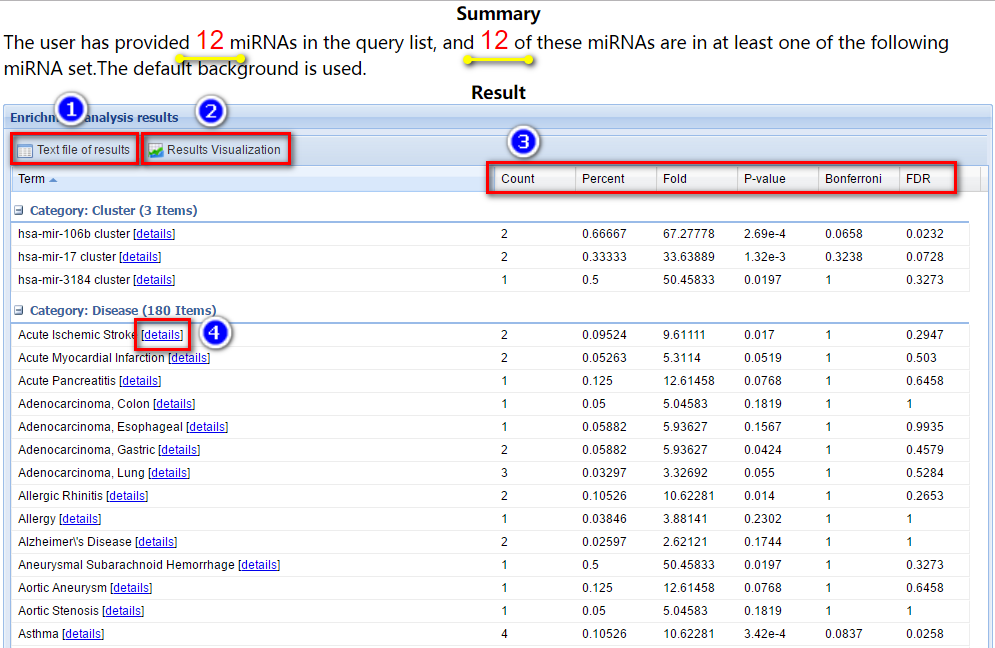
Before the tabular result, a brief summary of the user’s input is provided (as indicated by the yellow arrows in the figure below).
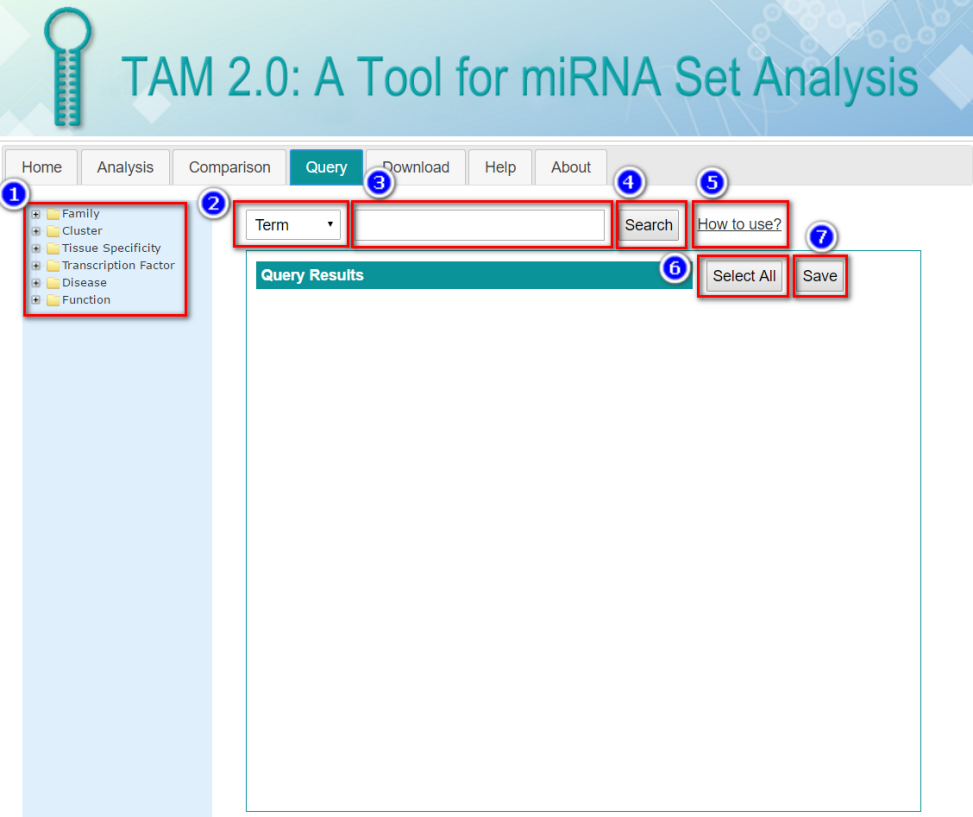
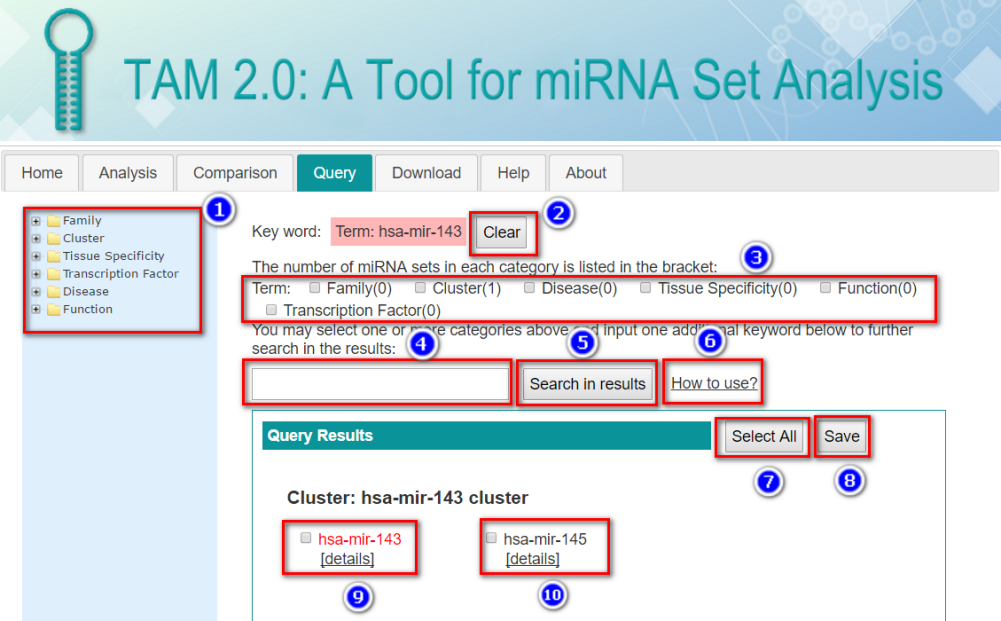
In this tabular result page, users can:
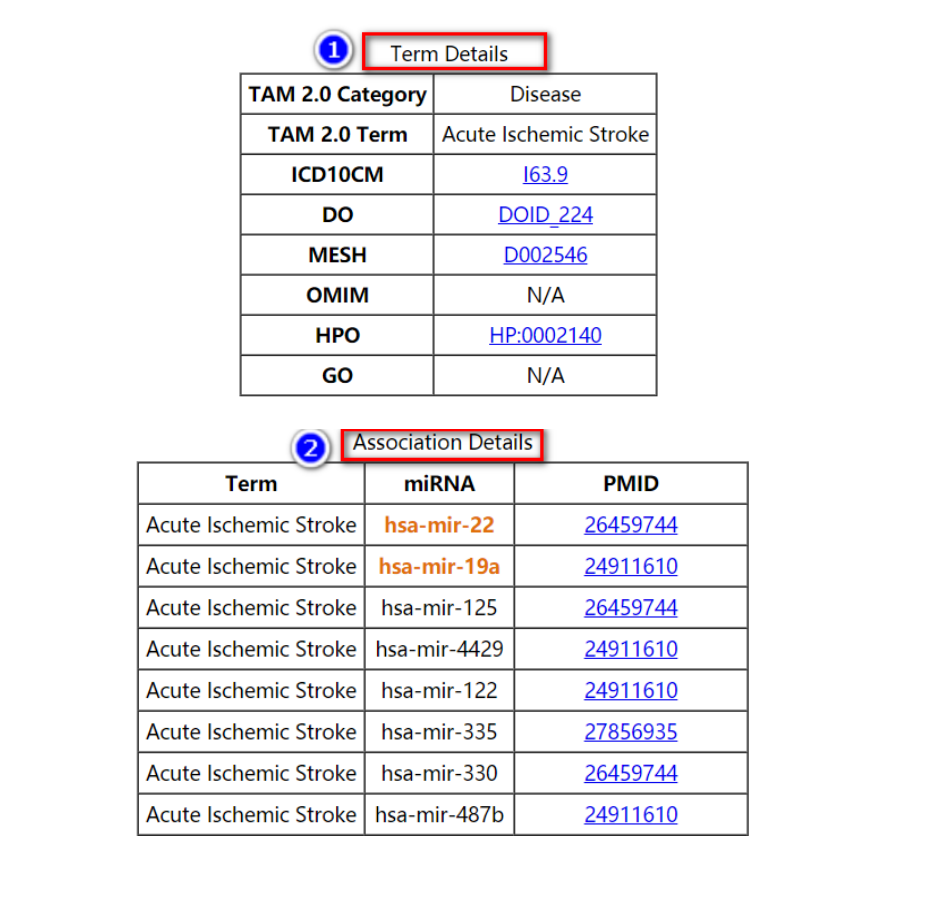
The Term Details table lists the cross-refence of the term to the standard terminology database including ICD-10-CM (ICD10CM), disease ontology (DO), NCBI MeSH term (MESH), NCBI OMIM (OMIM), human phenotype ontology (HPO) and gene ontology (GO). User can view the detail records in these databases by clicking the corresponding links.
While the Association Details table lists the PubMed ID (PMID) for each miRNA-set association. The miRNAs in input list are highlighted. User can view the detail PubMed reference records by clicking the corresponding links.
By clicking the “Result Visualization” in the tabular view of the analysis result, users are redirected to this page. User can generate plots illustrating the significant results, using the custom parameters:
.jpg)
In all, three types of plots are generated.
.png)
The number after the miRNA set term is the count of input miRNAs which are presented in each miRNA set.
.png)
This plot is similar to bar plot, whereas the size of bubble correlates with the count of input miRNAs which are presented in each miRNA set.
.png)
This heatmap depicts the holistic view of miRNA-miRNA set association. The color of each line in the heatmap correlated with the significance (P-value/Bonferroni/FDR, depends on the plot parameter selected above) of the association with each term.
Note that this page accepts human miRNA genes (pre-miRNAs) as its input. If the mature miRNA names (e.g. hsa-miR-140-5p) are provided, it will be collapsed into the corresponding miRNA gene (hsa-mir-140). If the exact name of the duplicated miRNA gene is not specified (e.g. hsa-mir-24), it will be mapped to all of the duplicated miRNA genes (hsa-mir-24-1 and hsa-mir-24-2).
User can generate plots illustrating the significant results, by simply 1) set the range of top terms to be displayed and 2) Generate the plots by clicking the Plot button.

In all, two types of plots are generated.
.png)
This bar plot illustrates the correlations (similarities) between user-provided de-regulated miRNAs and the de-regulated miRNAs in other disease conditions
.png)
This heatmap depicts the holistic view of miRNA de-regulation in the CUARATED DISEASE CONDITIONS. The palette correlates with the association score (de-regulation score, corrected for the specificity of miRNA de-regulation). The down-miRNAs are indicated by green boxes while the up-miRNAs are indicated by red boxes.

TAM 2.0 is the updated web server of our previously published miRNA set enrichment analysis tool, TAM (http://www.cuilab.cn/tam/) in 2010. Through manual curation of over 9,000 papers, a more than two-fold growth of reference miRNA sets has been achieved in comparison with previous TAM. We grouped miRNAs into six categories of miRNA sets: miRNA-family sets, miRNA cluster sets, miRNA-disease, miRNA-function sets, miRNA-TF sets and tissue specificity sets. Compared with the previous version of TAM, new functions for miRNA set query and result visualization are also enabled in the TAM 2.0. In all, TAM 2.0 provides a tool to mine the functional and disease implication behind miRNAs of interests.
This website has been tested by using Chrome, Microsoft Edge and Firefox browsers. Microsoft IE may not work well.
Here is a quick start tutorial:
Before you start: Please note, this page works for one list of interested miRNAs. If you simply wish to query the miRNA sets by a keyword (e.g. miRNA name), click here. If you have some up- and down-regulated miRNAs and are interested in the positive/negative correlations between your miRNAs with the de-regulated miRNA in other disease, you could also try the “comparison” tool.
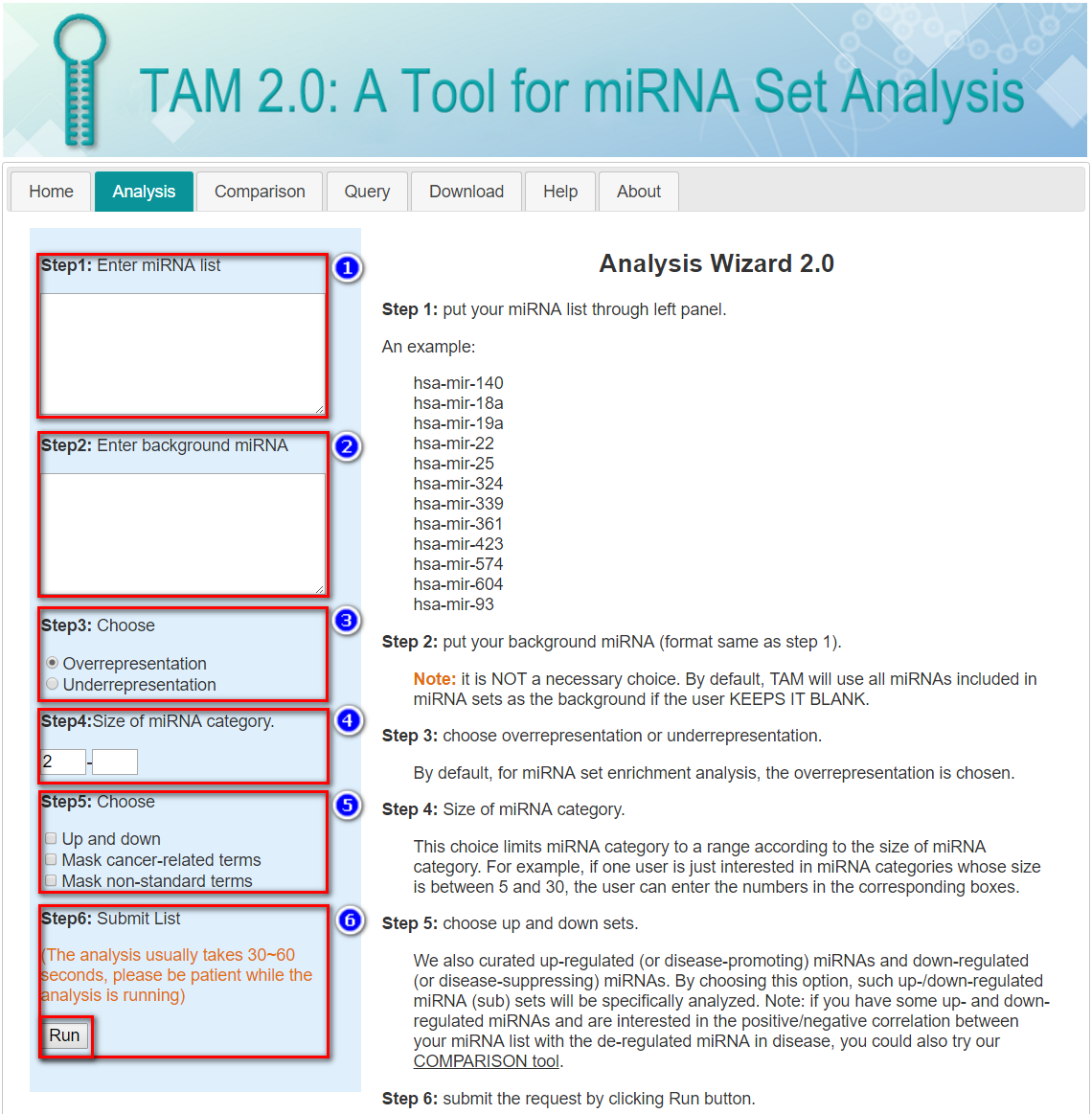
Step 1: Input your miRNA list through left panel.
Note: This page accepts human miRNA genes (pre-miRNAs) as its input, so the hsa-mir- prefix is assumed and the duplicated miRNA genes (e.g. with -1 or -2 suffix) would be specified.
An example:
If the mature miRNA names (e.g. hsa-miR-140-5p) are provided, it will be collapsed into the corresponding miRNA gene (hsa-mir-140). If the exact name of the duplicated miRNA gene is not specified (e.g. hsa-mir-24), it will be mapped to all of the duplicated miRNA genes (hsa-mir-24-1 and hsa-mir-24-2).
Step 2: Input your background miRNA (format same as step 1).
Note: It is NOT a necessary choice. By default, TAM will use all miRNAs included in the curated miRNA sets as the background if the user KEEPS IT BLANK.
Step 3: Choose overrepresentation or underrepresentation.
By default, for miRNA set enrichment analysis, the overrepresentation is chosen.
Step 4: Limit the size of miRNA set.
Step 5: Choose additional tag options.
Up and down: We also curated up-regulated (or disease-promoting) miRNAs and down-regulated (or disease-suppressing) miRNAs. By choosing this option, such up-/down-regulated miRNA (sub-) miRNA sets will be specifically analyzed.
Mask cancer-related terms: Because many experiments investigate cancer-related miRNAs, the cancer-related miRNA sets constitute a major part of our curated data. If you are not interested in cancer, select this option, and the cancer-related miRNA sets will be excluded from the analysis.
Mask non-standard terms: Some of our miRNA set terms cannot be mapped to the standard terminology databases (e.g. ICD-10-CM and Gene Ontology). If this option is chosen, these miRNA sets will be excluded from the analysis.
Step 6: Submit the request by clicking Run button.
TAM 2.0 results from collaboration between groups from Hebei University of Technology and Peking University Health Science Center. We try to understand life science using computing.
TAM 2.0 tool is free only for academic usage. For commercial useage, please contact the following PIs.
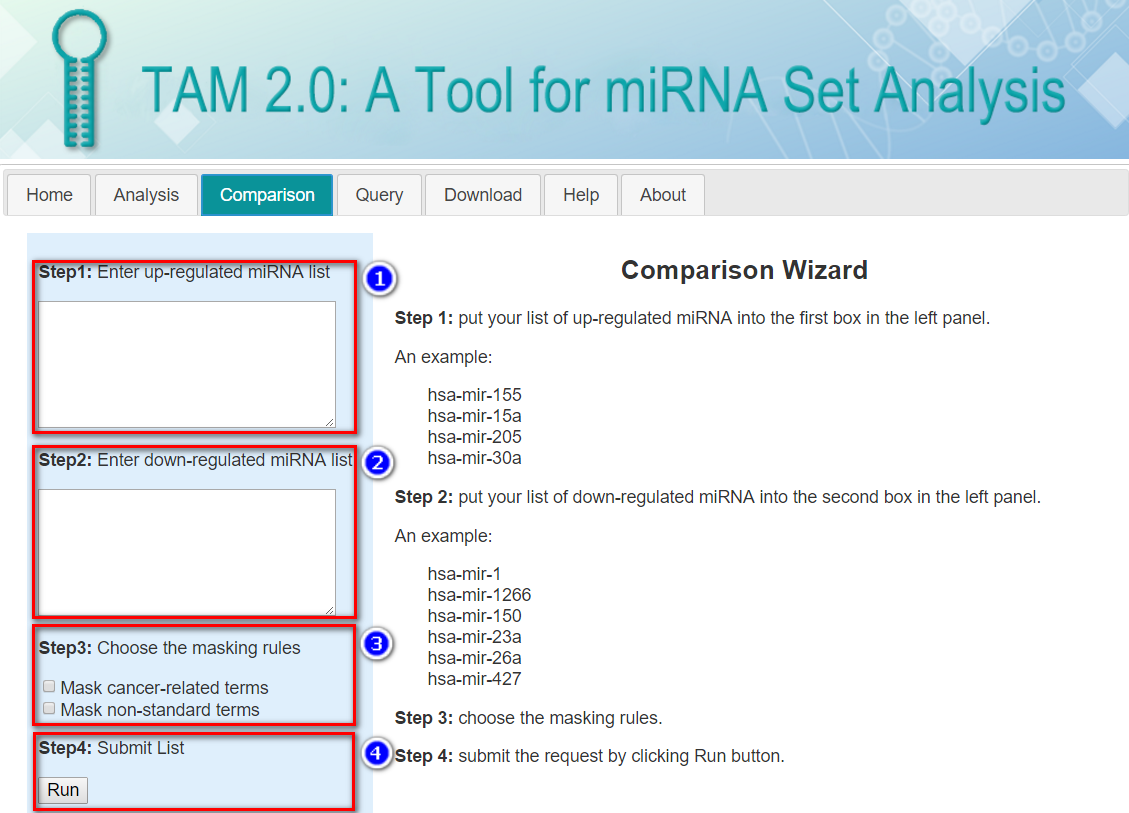
Group PIs:Before you start: Please note, this page works for two list of miRNAs (one list of up-miRNAs and one list of down-miRNAs). If you simply wish to query the miRNA sets by a keyword (e.g. miRNA name), click here. If you have only one list of interested miRNAs to be analyzed, please use the “analysis” tool instead.
Step 1: Input your list of up-regulated miRNAs into the first box in the left panel.
Note: This page accepts human miRNA genes (pre-miRNAs) as its input, so the hsa-mir- prefix is assumed and the duplicated miRNA genes (e.g. with -1 or -2 suffix) would be specified.
An example:
If the mature miRNA names (e.g. hsa-miR-155-3p) are provided, it will be collapsed into the corresponding miRNA gene (hsa-mir-155). If the exact name of the duplicated miRNA gene is not specified (e.g. hsa-mir-30c), it will be mapped to all of the duplicated miRNA genes (hsa-mir-30c-1 and hsa-mir-30c-2).
Step 2: Input your list of down-regulated miRNAs into the second box in the left panel.
An example:
Step 3: Choose additional tag options.
Mask cancer-related terms: Because many experiments investigate cancer-related miRNAs, the cancer-related miRNA sets constitute a major part of our curated data. If you are not interested in cancer, select this option, and the cancer-related miRNA sets will be excluded from the analysis.
Mask non-standard terms: Some of our miRNA set terms cannot be mapped to the standard terminology databases (e.g. ICD-10-CM and Gene Ontology). It this option is chosen, these miRNA sets will be excluded from the analysis.
Step 4: Submit the request by clicking Run button.
Citation: Jianwei Li, Xiaofen Han, Yanping Wan, Shan Zhang, Yingshu Zhao, Rui Fan, Qinghua Cui, and Yuan Zhou. TAM 2.0: tool for microRNA set analysis. Nucleic Acids Research, Volume 46, Issue W1, 2 July 2018, Pages:W180–W185.
Ming Lu, Bing Shi, Juan Wang, Qun Cao and Qinghua Cui. TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics 2010, 11:41